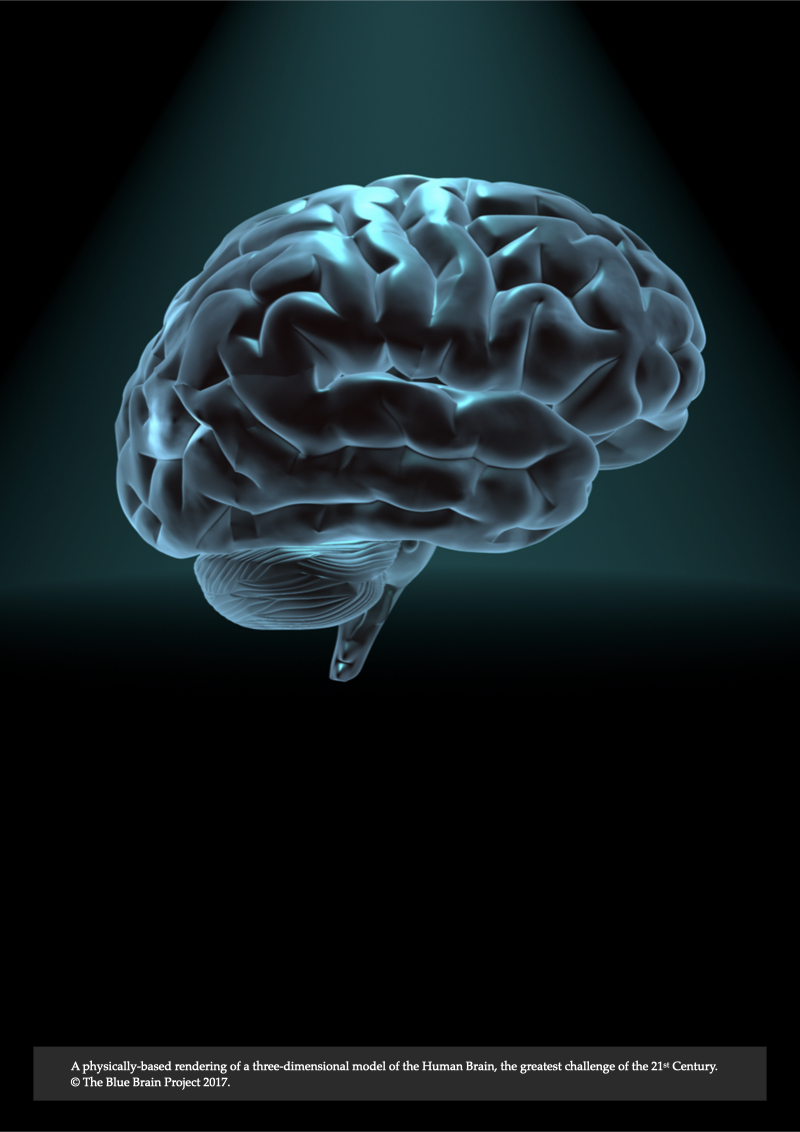
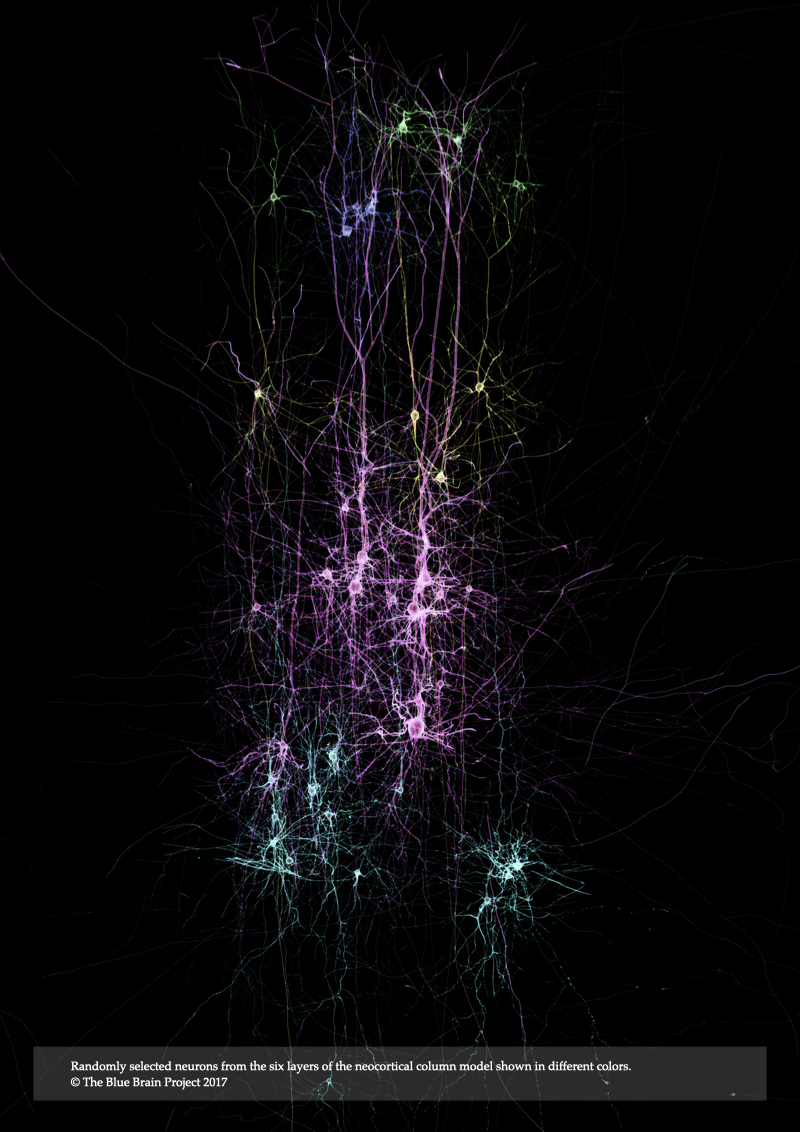
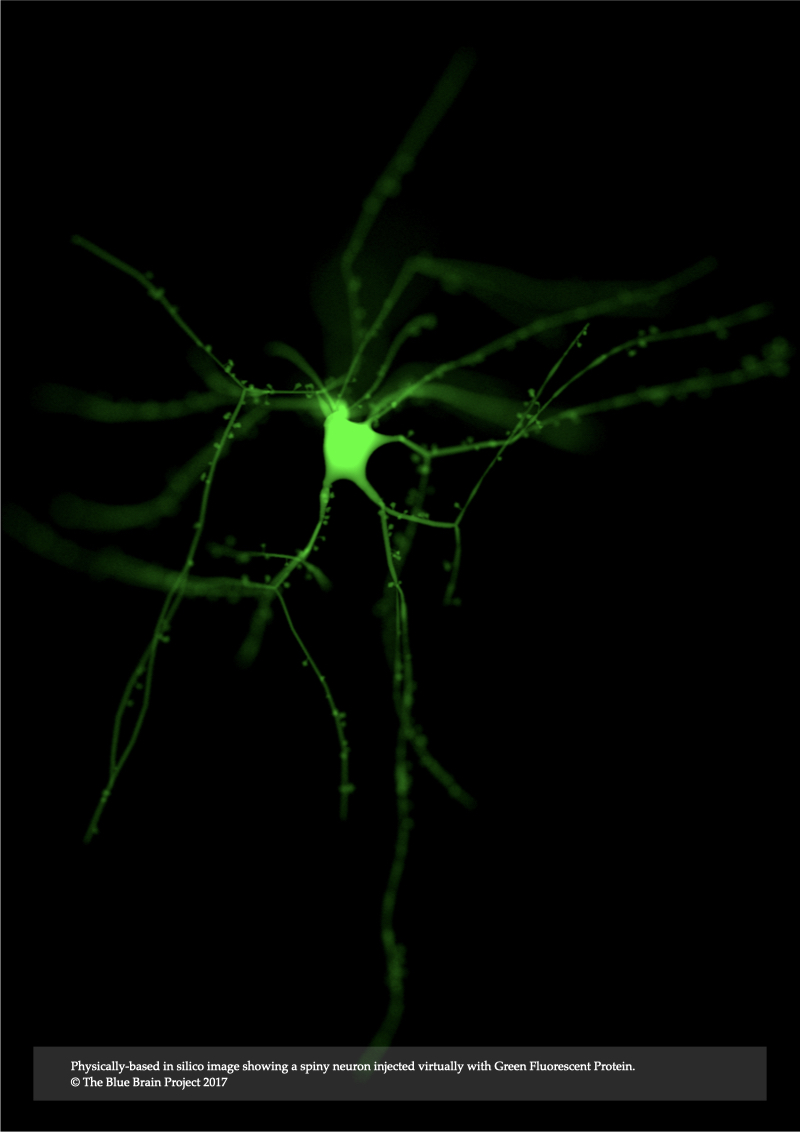
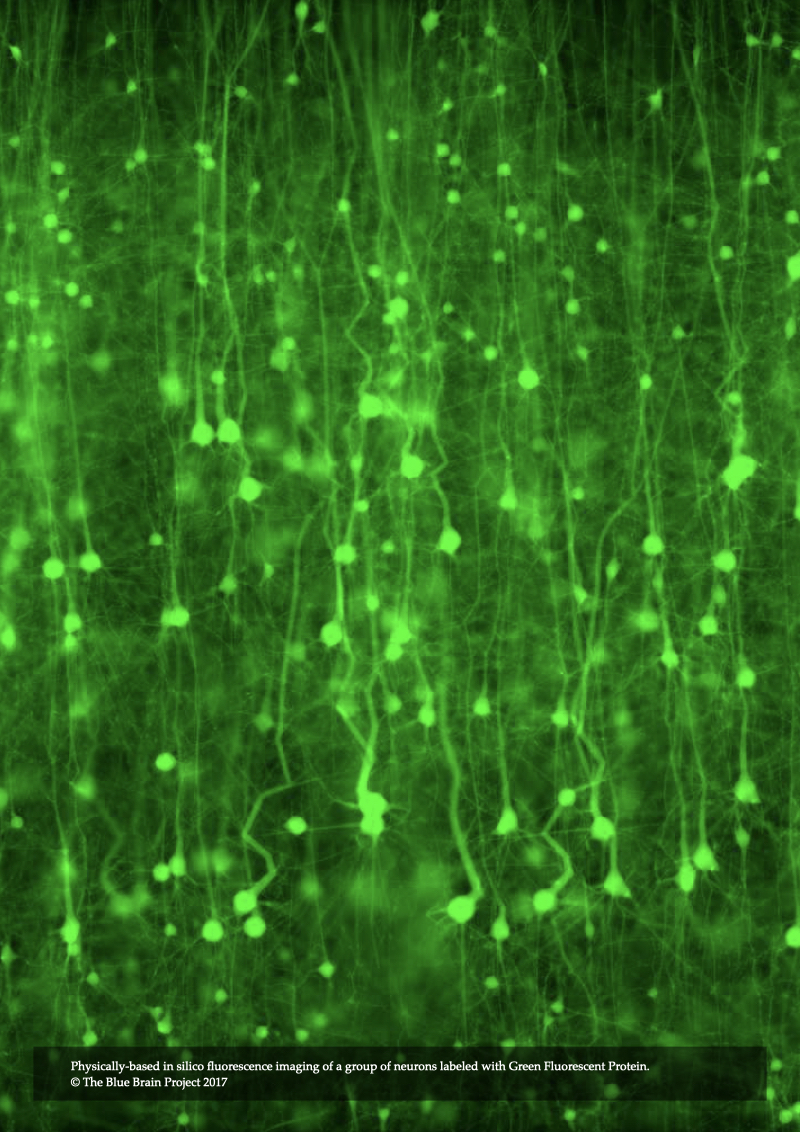
Record
- Title
- In silico brain imaging: physically-plausible methods for visualizing neocortical microcircuitry
- Doctoral School
- Neuroscience Doctoral School (EDNE)
Blue Brain Project (BBP)
Ecole Polytechnique Federale de Lausanne (EPFL)
- Academic Graduation Year
- 2017
- Examiners
-
Friedhelm Christoph Hummel, Jury President
Ivo F. Sbalzarini, External Examiner
Markus Axer, External Examiner
Dimitri Van De Ville, Internal Examiner
Henry Markram, Thesis Main Advisor
Felix Schürmann, Thesis Co-advisor
- Funding
- The research presented in this thesis was supported in part by the Blue Brain Project (BBP), the Swiss National Science Foundation under Grant 200020-129525 and the King Abdullah University of Science and Technology (KAUST) through the KAUST – EPFL alliance for Neuro-Inspired High Performance Computing.
Abstract
The recent years have witnessed growing interest in the power of scientific visualization for simulation-based neuroscience. The research presented in this thesis develops methods to generate physically-realistic visualizations of neocortical models reconstructed by the Blue Brain Project, a pioneering endeavor that uses a simulation-based approach to build large-scale, biologically-detailed digital reconstructions of neocortical microcircuitry. The project has established a domain-specific framework to visualize hybrid representations of these neocortical models, which it uses to evaluate their structure, composition and dynamics. The current framework offers naïve, optical emission-only models that oversimplify light-tissue interaction and ignore other computationally expensive phenomena such as absorption, scattering and fluorescence. The methods presented in this dissertation overcome these limitations, making it possible to visualize neocortical models on a physically-plausible basis, taking into account the intrinsic optical properties of cortical tissue and the spectroscopic characteristics of its fluorescent structures. This requires rigorous optical models that accurately simulate light interaction with cortical tissue, and accurate volumetric models of the tissue itself which reflect its optical properties during simulation. We present an efficient framework for creating high fidelity large-scale volumetric models of neocortical microcircuitry that govern the way in which light is distributed in cortical tissue. These models are reconstructed in two steps: first, we build piecewise watertight mesh models of neocortical neurons from their morphological skeletons. Then, we apply solid voxelization to the meshes to build volumetric models. We also present two novel optical models for simulating light interaction with fluorescent structures in low- and highly-scattering volumes. These optical models are integrated into a high level framework that simulates the imaging pipelines of transmitted light brightfield, wide-field epi-fluorescence and light-sheet fluorescence microscopes. It is based on the principles of geometric optics and Monte Carlo ray tracing. Afterwards, we exploit this framework to render in silico realistic microscopic images resembling those created by the actual instruments. In conclusion, we generalize the concept of physically-based simulation of brain imaging modalities and review the computational models required to simulate fMRI.
Preface
Chapter 1, Introduction, gives a gentle overview of the topic and discusses the substantial role of physically-based visualization in the context of simulation-based neuroscience. The limitations of previous visualization methods are discussed, the motivations that have initiated the spark of this work are elaborated and finally our major contributions are summarized.
In Chapter 2, Realistic Neocortical Tissue Modeling for In Silico Imaging, we present a systematic approach for building realistic large scale volumetric models of the neocortical circuity from the morphological representations of the neurons. Those model account for light interaction with the different structures of the tissue.
The optical models presented in this thesis require certain level of knowledge of the theory of light transport in volumetric media. Chapter 3, Fundamentals of Light Interaction with Brain Tissue, lays down the basic fundamentals of light transport that are necessary to allow the reader to understand the derivation of the fluorescence models presented in the following chapters.
The light transport equation presented in Chapter 3 represents a more rigorous model than the emission-only model that is commonly used in interactive rendering applications. It accounts for absorption and elastic scattering, but it ignores light interaction with fluorescent volumes. Chapter 4, Modeling Light Interaction with Low Scattering Fluorescent Volumes, presents a novel optical model that extends the monochromatic rendering integral taking inelastic scattering, or fluorescence, into account. This extended model is capable of rendering low-scattering fluorescent neocortical data. The model is expressed in terms of the spectroscopic properties of fluorescent dyes and their concentration when injected into the intracellular space of the neurons.
The fluorescence model presented in Chapter 4 is limited only to low scattering fluorescent volumes. It can be utilized to render fluorescent images of clarified brain models assuming the removal of all the lipid content that is characterized with high turbidity, but it fails otherwise. In Chapter 5, Modeling Light Interaction with Highly Scattering Fluorescent Volumes, we present a further extension to account for light transfer in highly scattering fluorescent volumes, taking into account the spectroscopic properties of fluorescent dyes in addition to the optical properties of the tissue itself. The spectral performance of this optical model is also validated against normalized emission spectra of multiple fluorescent dyes. The results of the model are demonstrated with rendering three neocortical models tagged with fluorescent solutions that have low-, medium- and high scattering properties.
In Chapter 6, In Silico Transmitted Light Brightfield Microscopy, we present a computational model of brightfield microscopy including its illumination and acquisitions units in addition to light interaction with a virtual specimen that is filled with highly attenuating dye to improve the contrast in the final image. This microscopic model is exploited afterwards to simulate the process of imaging brain tissue stained with Golgi’s method using a group of few pyramidal cells filled virtually with an opaque dye that has optical properties similar to those of a typical Golgi’s solution.
In Chapter 7, In Silico Fluorescence Microscopy, the rendering integrals of fluorescence that were derived in Chapter 4 and Chapter 5 are integrated into our microscopy simulation framework to introduce a novel model for epi-fluorescence microscope. This model is applied to visualize annotated models of neocortical circuitry virtually injected with various fluorescent dyes that are of significance in neurobiology. The microscopic model is also employed to perform several in silico experiments to investigate the visual re- sponse of the same specimen model when excited with a laser source at different wavelengths.
In Chapter 8, In Silico Light Sheet Fluorescence Microscopy, the illumination and acquisition models of the epi-fluorescence microscope are extended to introduced a novel model of structured illumination microscopy. This model allows rendering fluorescent images of the neocortical tissue with high axial resolution and less blurring artifacts. The model is then used to carry out a set of in silico experiments to visualize annotated tissue models when illuminated with different light sheet thicknesses. Moreover, it is used to simulate brainbow experiments when the tissue is labeled with multiple fluorescent dyes having different colors at the same time.
The introduction of functional Magnetic Resonance Imaging (fMRI) has pioneered neuroscience research to address the mapping of the functional aspects of the human brain. Since its inception, several models and simulation studies for blood oxygen-level dependent (BOLD) fMRI were proposed and extensively discussed. Most of these research studies were focused on understanding the underlying mechanisms of the technique. The complexity of fMRI data challenged the development of common, precise and physiologically-plausible fMRI simulators, and consequently the data generation models in the majority of the current simulators are ad hoc. In Chapter 9, Computational Modeling & Simulation of fMRI, we present an extensive review of the modelling work and numerical simulation studies of BOLD fMRI that were covered in the literature to date. This chapter is not intended to present a simulator for fMRI; this is beyond the scope of this thesis, but rather, to generalize the concept of in silico imaging to a non-optical instrument.
Chapter 10, Conclusions, Perspectives and Future Work, summarizes the major results of the thesis and discusses several extensions of our microscopy simulation framework and its applications to reconstruct and visualize other kinds of data including vasculature, glial cells and also different brain regions such as the hippocampus.
Downloads
- Individual Chapters
- Front Matter
- Chapter 1: Introduction: In Silico Brain Imaging
- Chapter 2: Realistic Neocortical Tissue Modeling for In Silico Imaging
- Chapter 3: Fundamentals of Light Interaction with Brain Tissue
- Chapter 4: Modeling Light Interaction with Low Scattering Fluorescent Volumes
- Chapter 5: Modeling Light Interaction with Highly Scattering Fluorescent Volumes
- Chapter 6: In Silico Transmitted Light Brightfield Microscopy
- Chapter 7: In Silico Fluorescence Microscopy
- Chapter 8: In Silico Light Sheet Fluorescence Microscopy
- Chapter 9: Computational Modeling & Simulation of fMRI
- Chapter 10: Conclusions, Perspectives and Future Work
- Appendix A: Software Guide
- Appendix B: PBRT Configuration Files
- Appendix C: Exemplars
- Appendix D: Spectra
- Bibliography