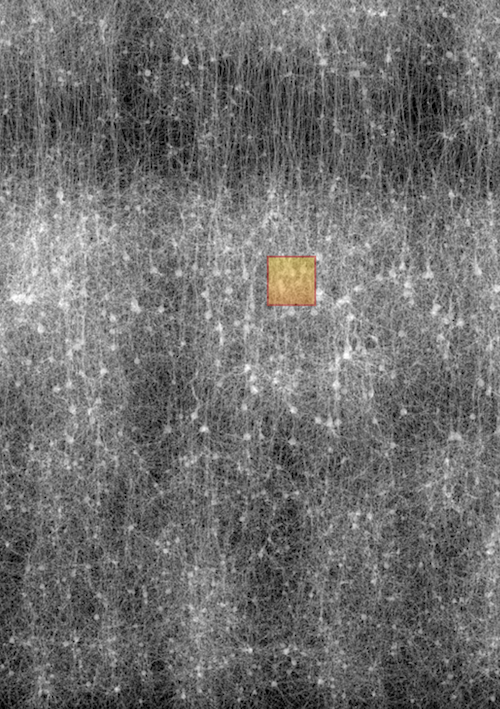
Research & Development
Mesh Reconstruction

Generation of high fidelity, two-manifold and watertight polygonal surface meshes
of neuroscientific models including spiny neurons, glial cells and blood vasculature
from high
resolution segmented electron microscopy stacks.
These polygonal meshes are used to create homogeneous tetrahedral or volumetric
meshes for
reaction-diffusion simulations, for example using the
STEPs
simulator, to understand the functional
mechanisms of
neuro-glio-vascular ensemble.
Neuronal Visualization
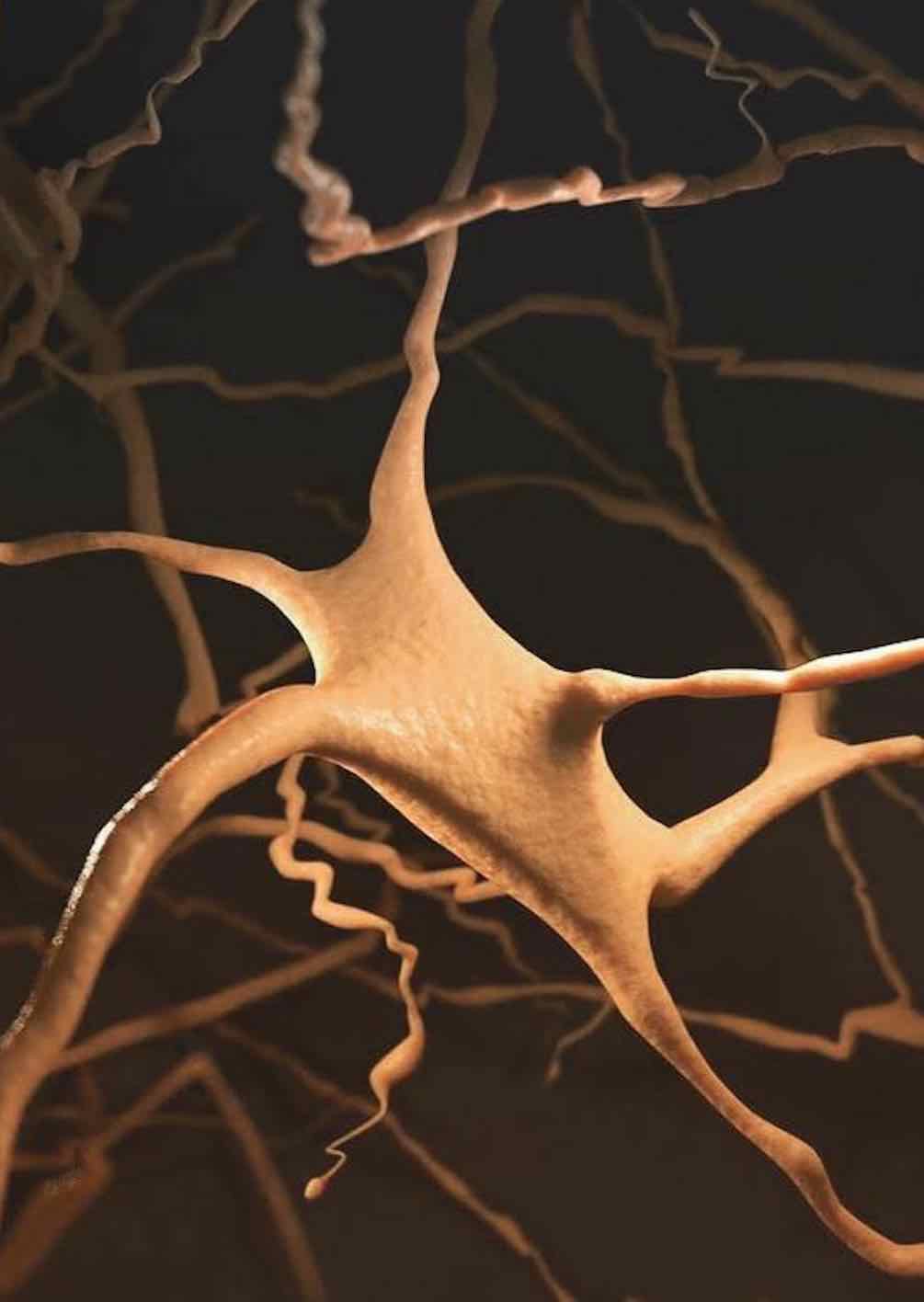
Developing NeuroMorphoVis;
an integrated, interactive, extensible and cross-platform framework for building,
visualizing and analyzing digital
reconstructions of neuronal morphology
skeletons
extracted from microscopy stacks. The framework is capable of detecting
and repairing several tracing artifacts, allowing the generation of high fidelity
surface meshes and high resolution
volumetric models for simulation and in silico studies. NeuroMorphoVis is based on
Blender and is freely available on
Github.
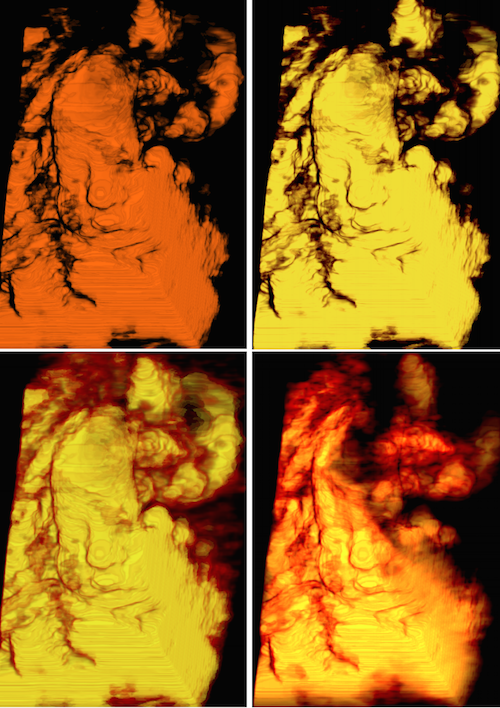
Brain Vasculature

Developing VessMorphoVis;
an integrated suite of toolboxes for interactive visualization and analysis of vast
brain vascular networks
represented by morphological
graphs
segmented originally from imaging or microscopy stacks.
The tool leverages the outstanding potentials of Blender, aiming to establish an
integrated, extensible
and domain-specific framework capable of interactive visualization, analysis,
repair, high-fidelity
meshing and high-quality rendering of vascular morphologies.
VessMorphoVis is based on Blender and is freely available on
Github.
Fluorescence Rendering
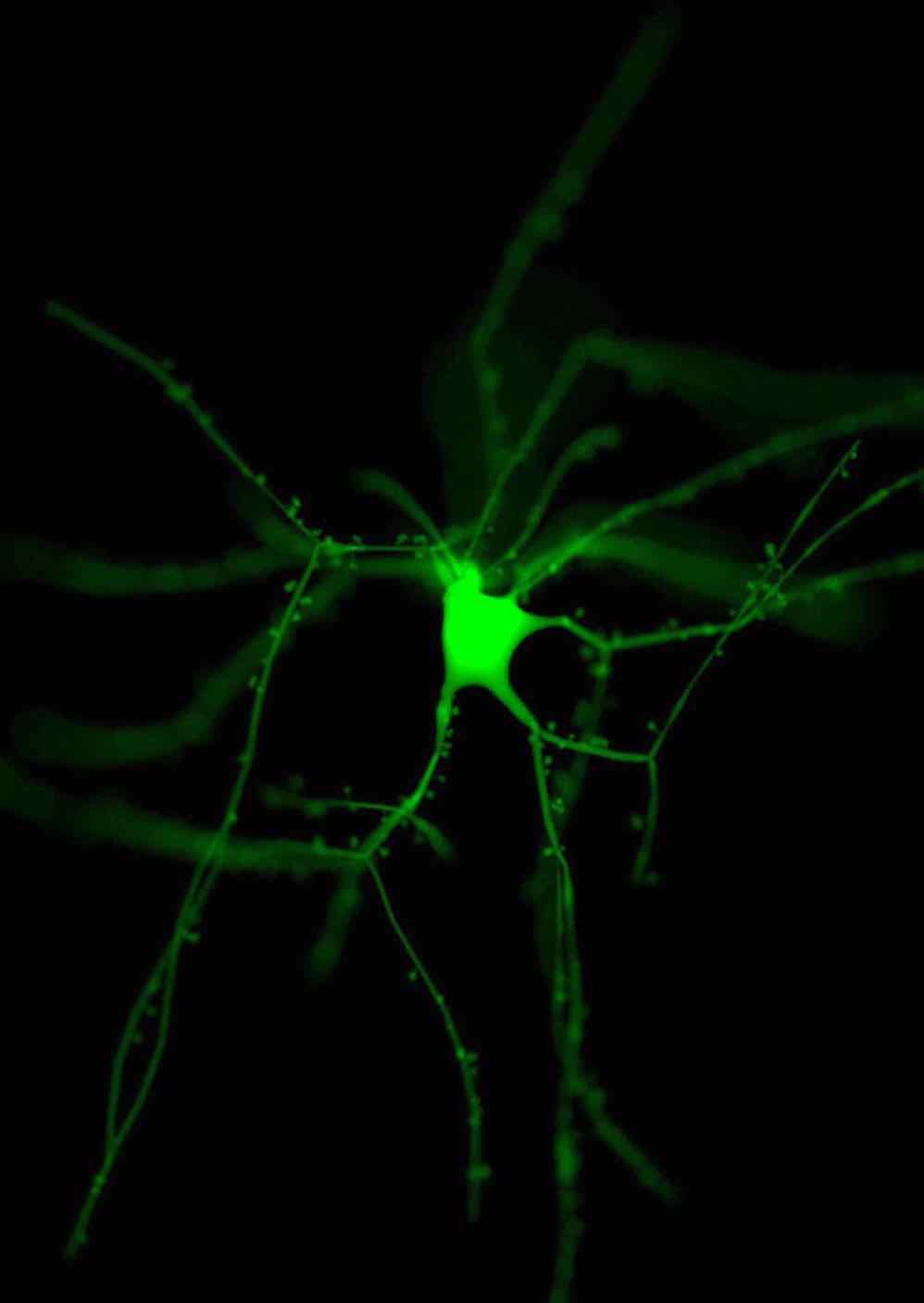
Development of efficient and unbiased physically-plausible optical models for
rendering fluorescent
structures in low- and highly-scattering turbid media. These models can be further
employed to
simulate the optical pipelines of several fluorescence-based optical microscopes,
such as the
epi-widefield or the light-sheet fluorescence microscopes, to digitally reconstruct
in silico
fluorescence imaging experiments. The models can also be used to simulate other
types of experiments
such as voltage sensitive dyes imaging and Calcium imaging.
Marwan Abdellah's Page
All Rights Reserved
Last updated on 01.09.2025